Gravitational Attraction
What would happen if two people out in space a few meters apart, abandoned by their spacecraft, decided to wait until gravity pulled them together? My initial thought was that …
In #programming
A while ago I wrote this little package called pyndamics which was a thin wrapper around the scipy odeint function. Since then I migrated it using nbdev to experiment with using jupyter notebooks as the basis for making packages, yielding pyndamics3. I added MCMC support for parameter fitting fairly early, using PyMC, but changed to emcee to have an all-python implementation. (This past month, after upgrading to Big Sur on my Mac, I've been struggling to even install PyMC3 and pystan due to dependencies but that is another story).
Recently, it struck me that the lmfit package might be a straightforward way to add some curve fitting that would work with simpler systems, and take less time than running MCMC. So, now pyndamics3 can use lmfit as a fitting backend. For example,
%pylab inline
from pyndamics3 import Simulation
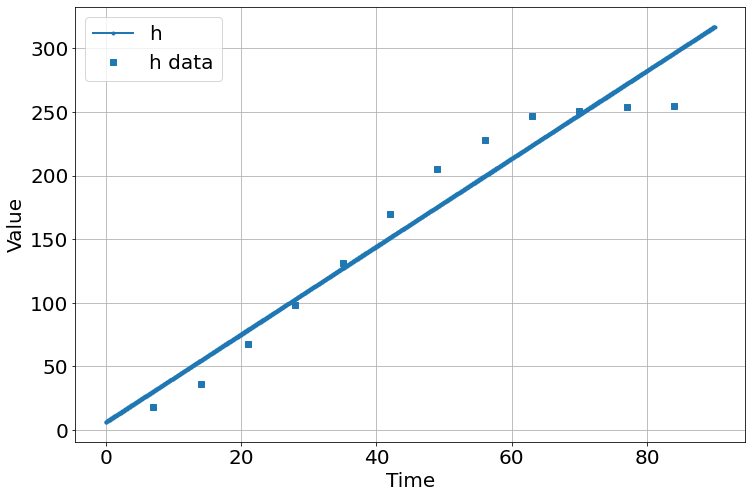
t=array([7,14,21,28,35,42,49,56,63,70,77,84],float)
h=array([17.93,36.36,67.76,98.10,131,169.5,205.5,228.3,247.1,250.5,253.8,254.5])
sim=Simulation()
sim.add("h'=a",1,plot=True)
sim.add_data(t=t,h=h,plot=True)
sim.params(a=1)
sim.run(0,90)
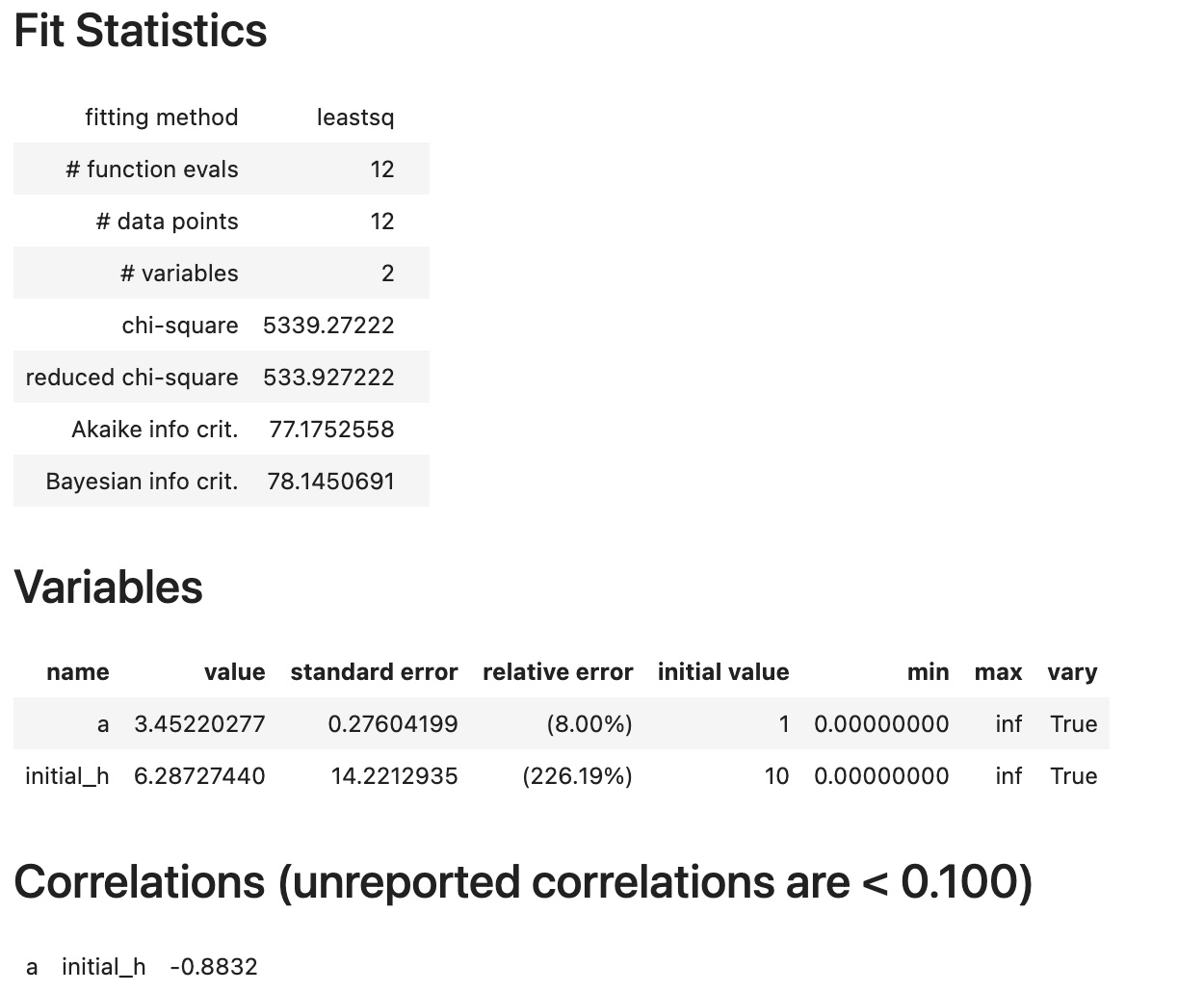
from pyndamics3.fit import fit,Parameter
results=fit(sim,
Parameter('a',value=1,min=0),
Parameter('initial_h',value=10,min=0))
results
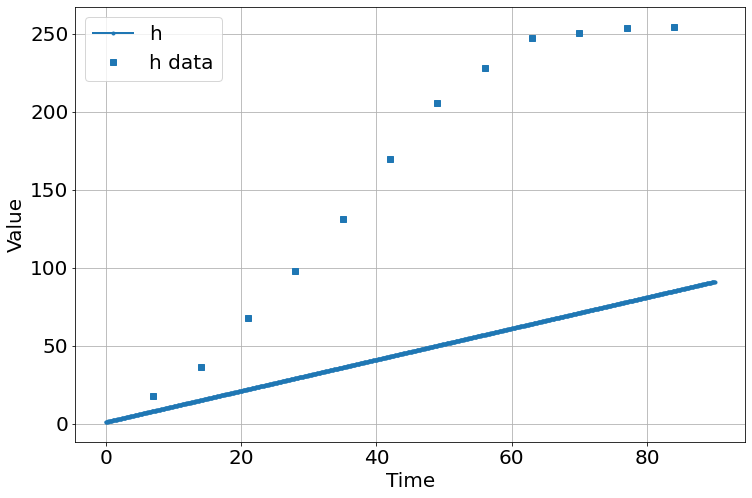
sim=Simulation()
sim.add("h'=a*h*(1-h/K)",1,plot=True)
sim.add_data(t=t,h=h,plot=True)
sim.params(a=2,K=100)
sim.run(0,90)
with
results=fit(sim,
Parameter('a',value=1,min=0.01,max=2),
Parameter('K',value=1,min=0,max=400),
Parameter('initial_h',value=10,min=0,max=50),method='powell')
report_fit(results)
sim.run(0,90)
(notice specifying the method).